Optimized LBM
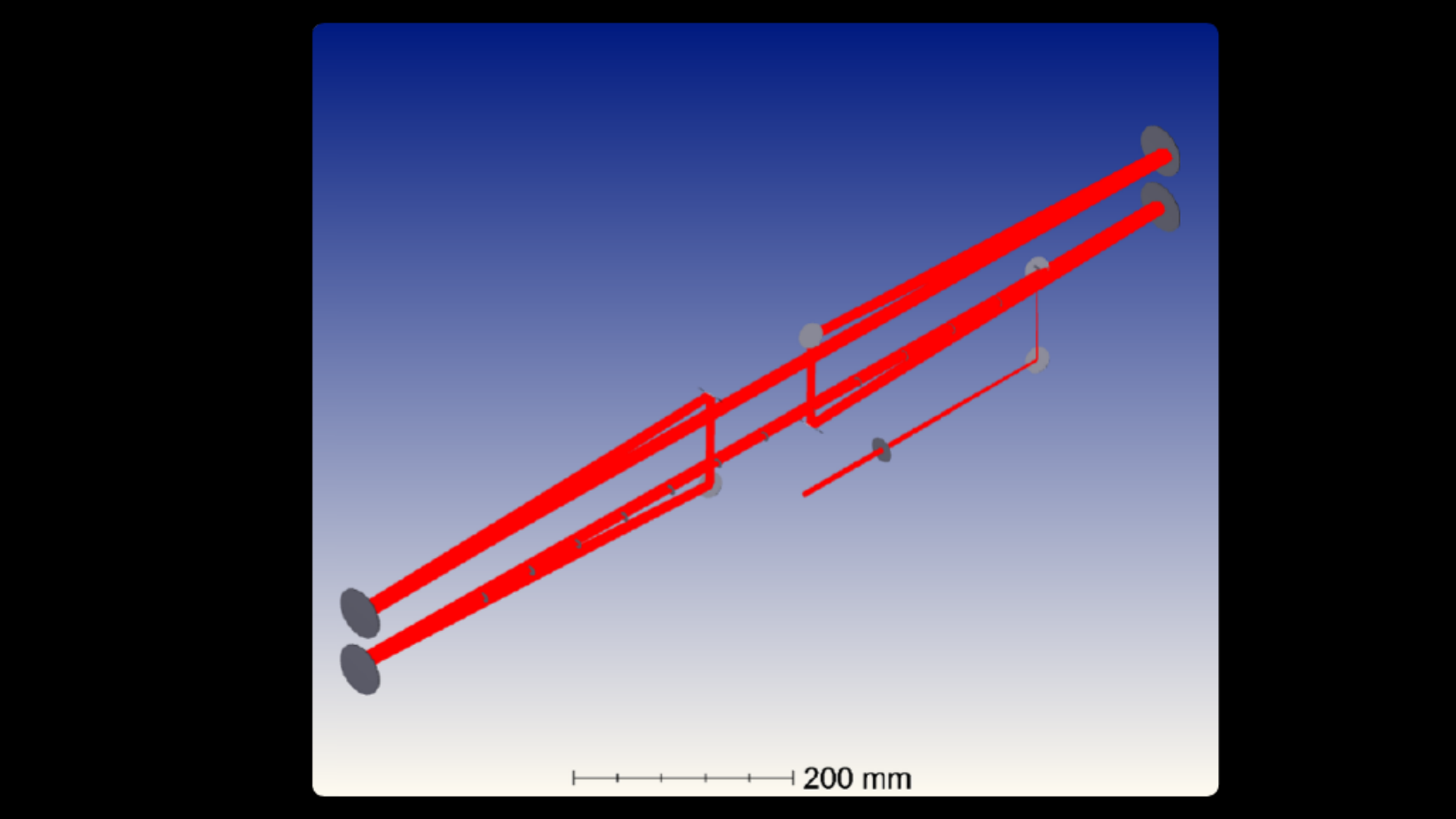
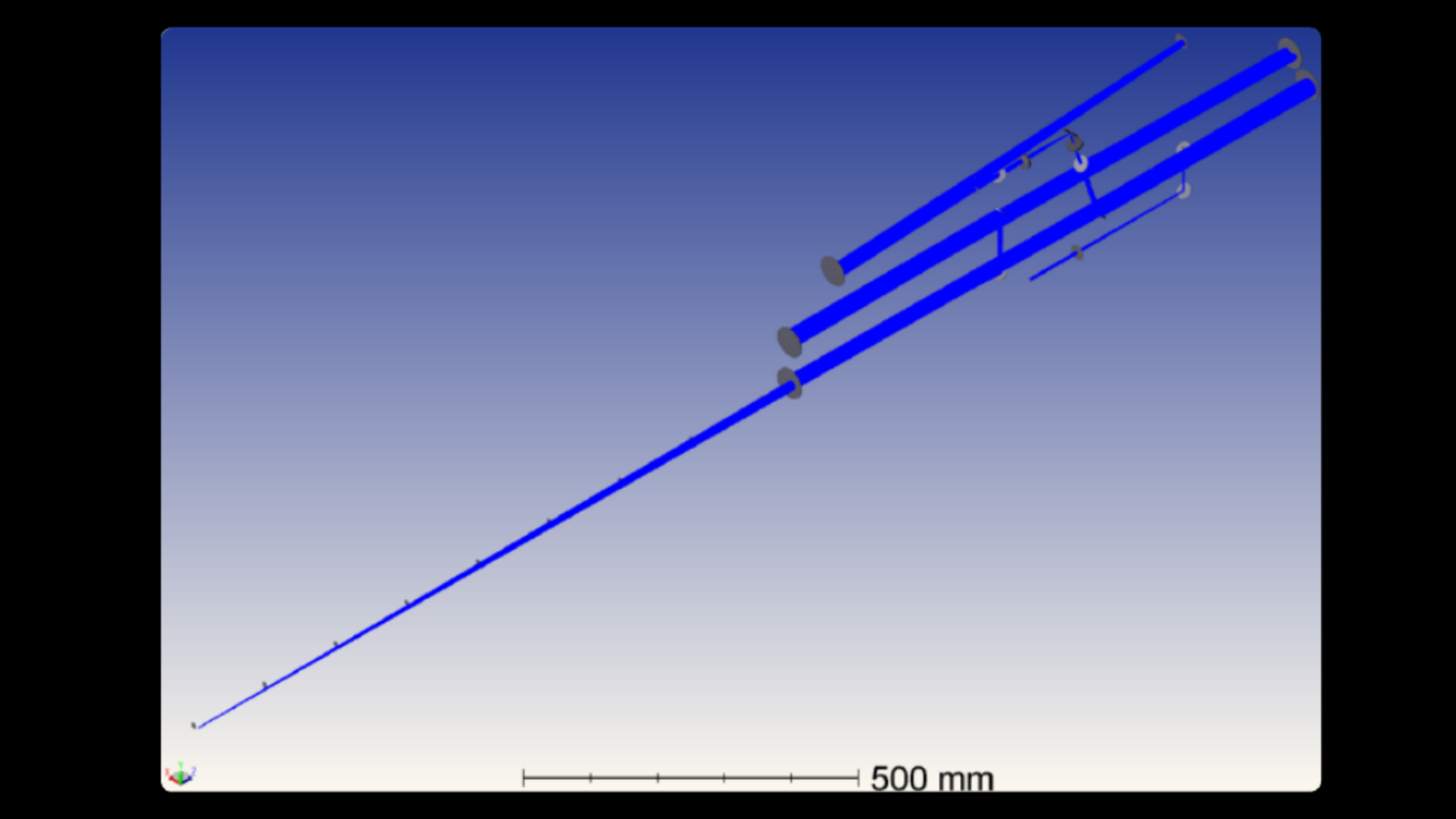
The Miller Brain Observatory has developed an optimized LBM module and paired it with an open-source microscope design, MIMMS (Modular In vivo Multiphoton Microscopy System). This refinement building upon the proof-of-principle LBM design [1] and achieves sub-cellular resolution by generating a column of light-beads from each high-energy pulse emitted by a low repetition laser source (5 MHz). The light-beads span multiple axial locations (up to 30 across 500 microns) within a sample volume, with each light-bead’s power optimized as a function of depth. Furthermore, LBM utilizes a single excitation pulse to optimally address each voxel. As such, volumetric recording to capture large fields of view is possible at the rate a single plane is recorded by conventional 2-photon microscopy. An in-depth overview of the technology is provided in the summary write-up.
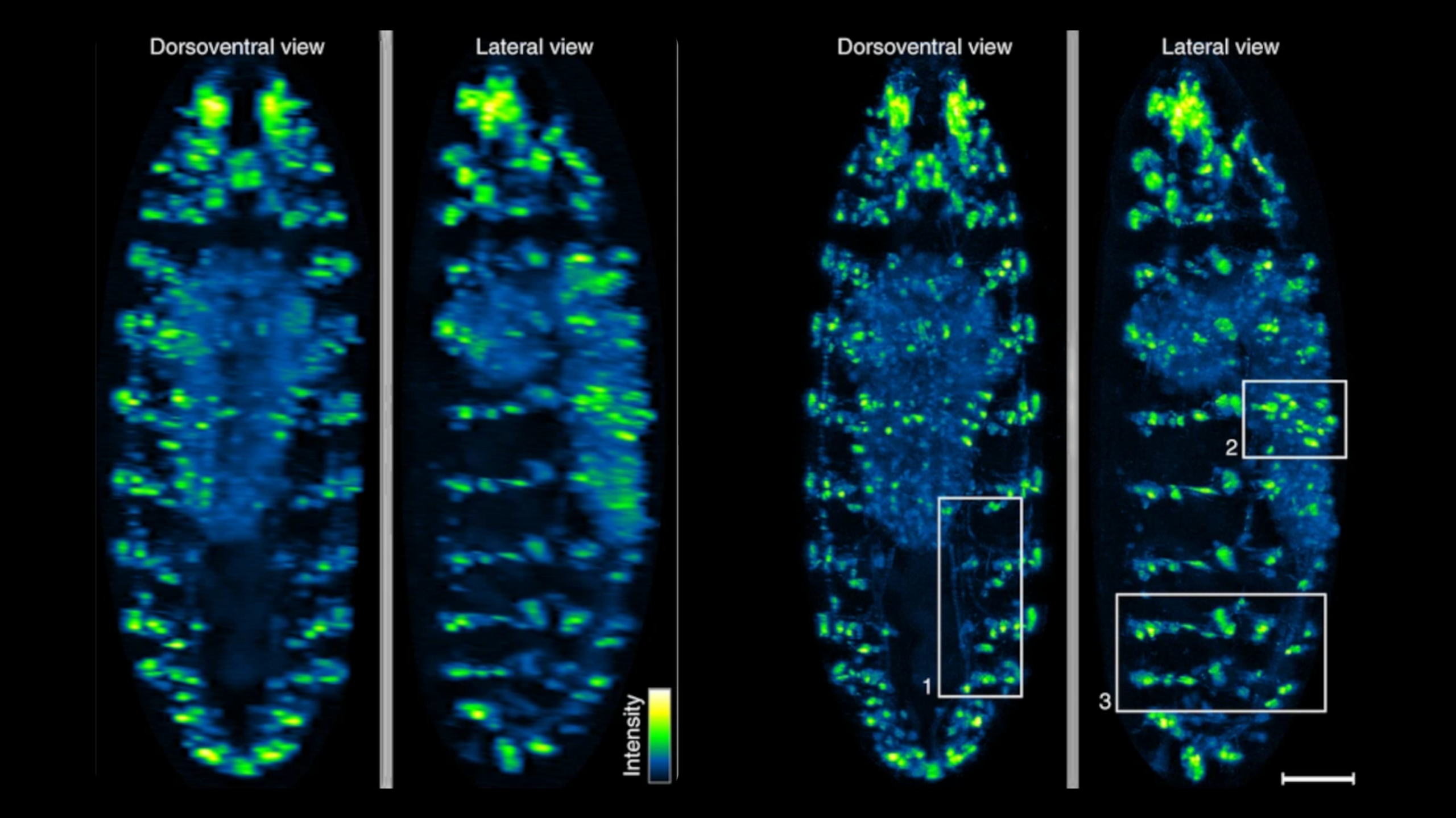
Biological applications unlocked by LBM include a recent study [2] which revealed an unbounded scaling of neuronal dimensionality with the number of neurons, which was only possible due to the simultaneously recording of activity from one million neurons in mice. Another recent example includes a study [3] showing how working memory is improved and consolidated with repetitive practice, for which 73,000 cortical neurons in mice were simultaneously observed as the animals learned and repeated their task.
High-Resolution LBM
The Miller Brain Observatory has developed a specialized high-resolution configuration that achieves sub-micron spatial resolution while maintaining volumetric imaging rates of up to 20 Hz.
This configuration preserves LBM’s core advantages—single-pulse voxel excitation and spatiotemporally optimized sampling—while tuning the system for enhanced optical resolution. As a result, users can resolve individual cellular compartments such as dendritic spines and axonal boutons across large cortical volumes.
This configuration is well-suited to capture whole-brain activity at high spatial and temporal resolution in invertebrate model organisms, and is poised to open new avenues to investigate the fine-scale dynamics of neural activity, including synaptic integration, compartment-specific plasticity, and the subcellular correlates of learning, memory, and behavior.
References
[1] Demas et al., Nature Methods (2021)
[2] Manley et al., Neuron (2024)
[3] Bellafard et al., Nature (2024)
IsoView
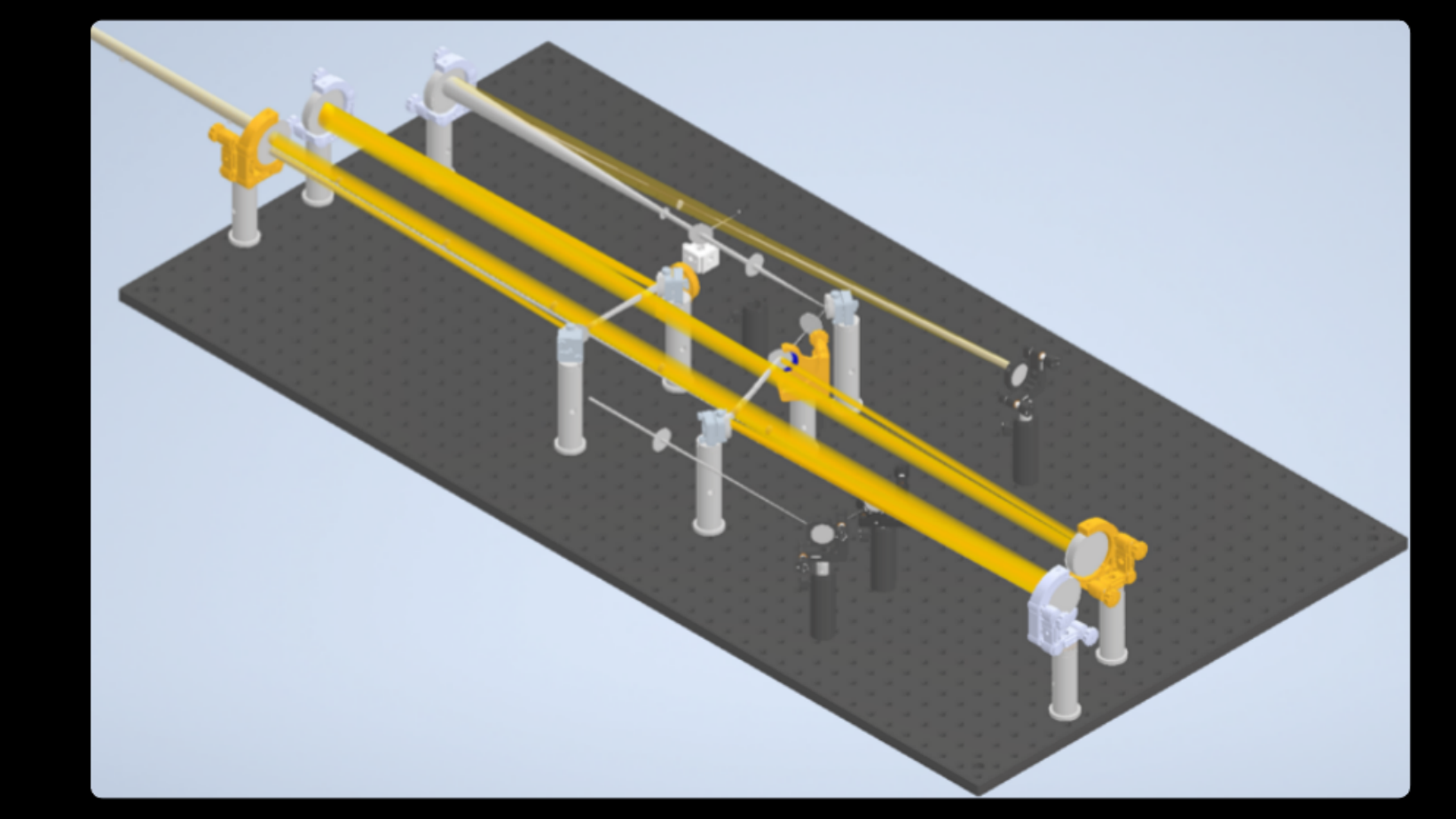
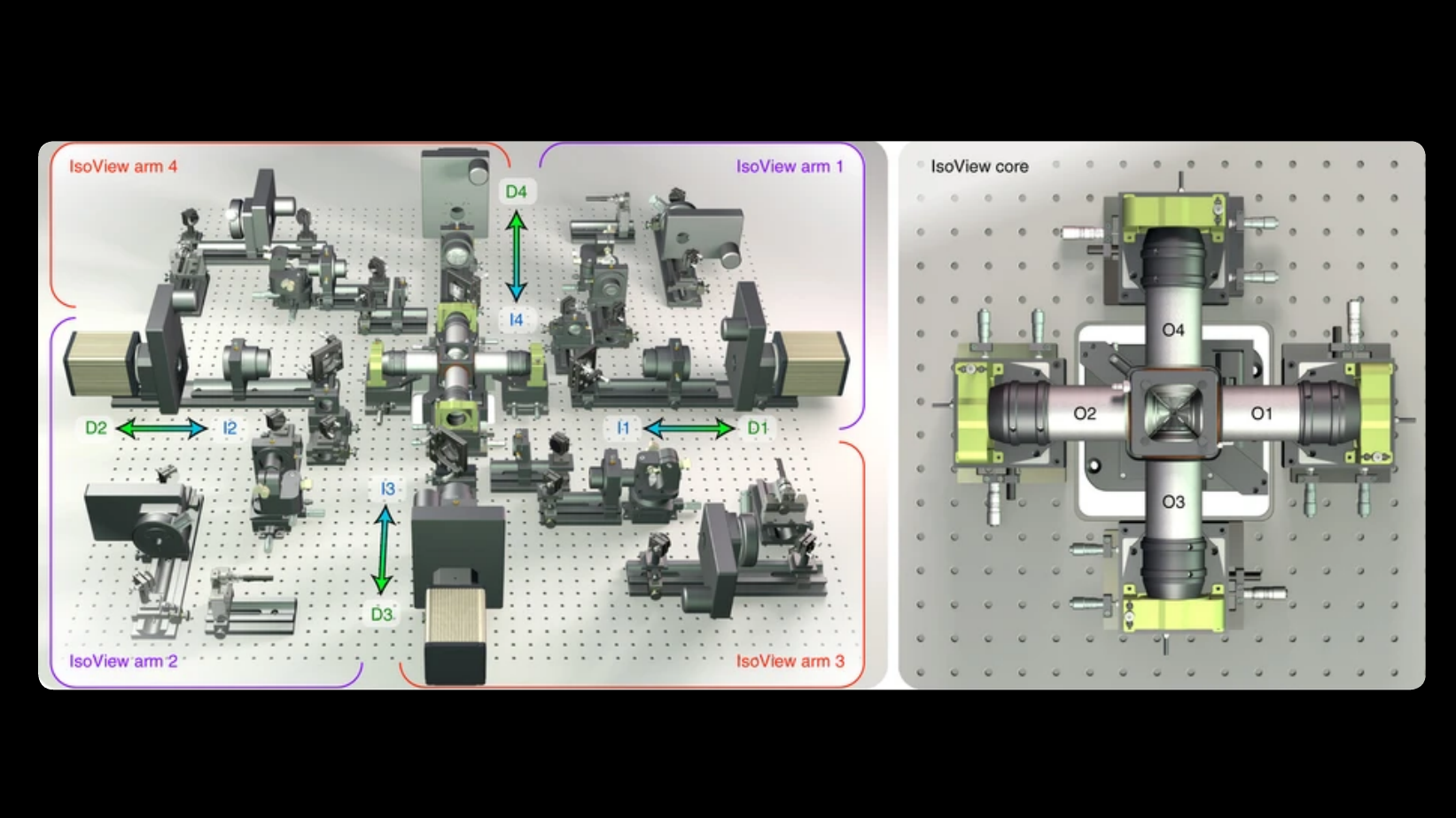
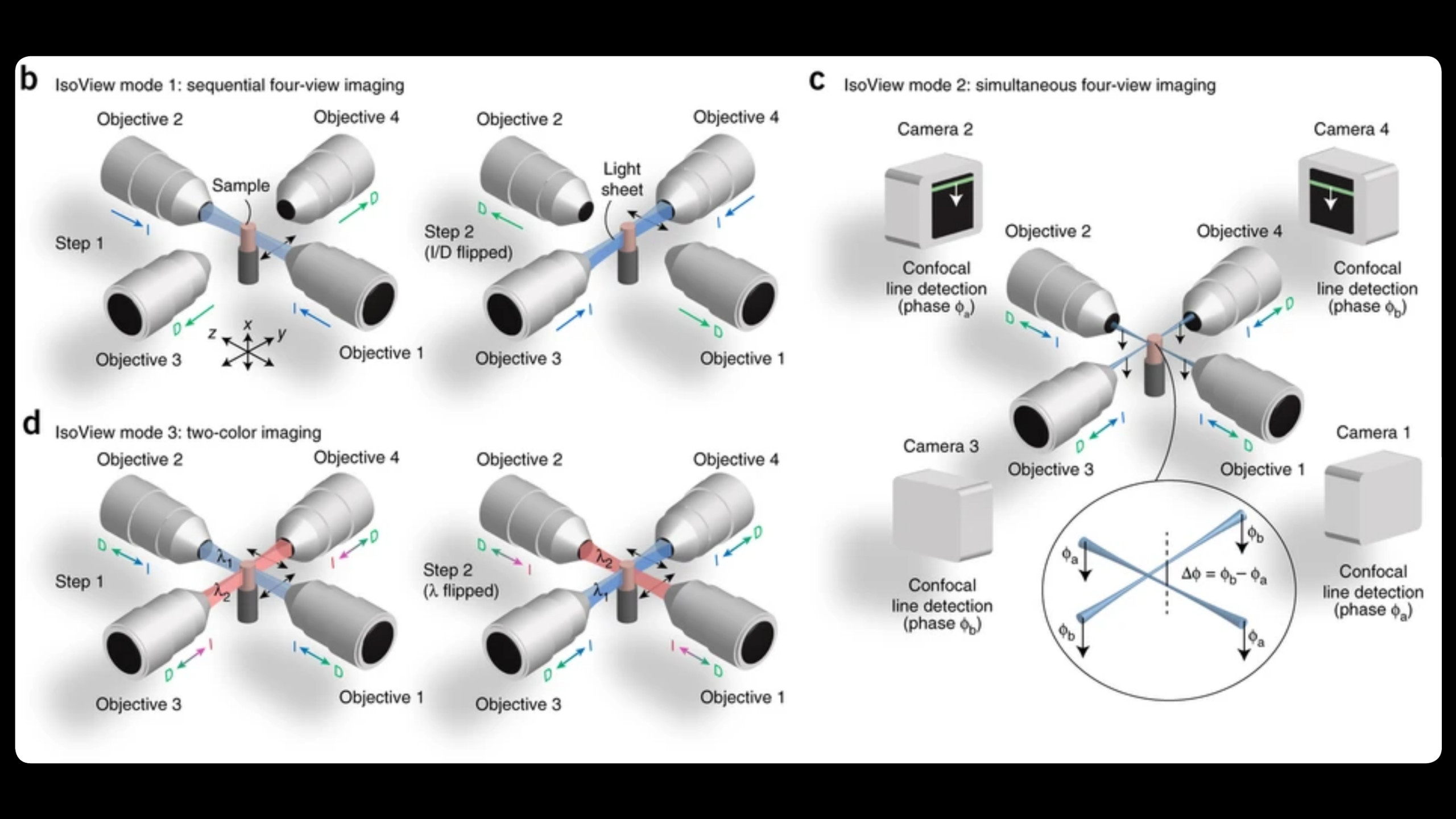
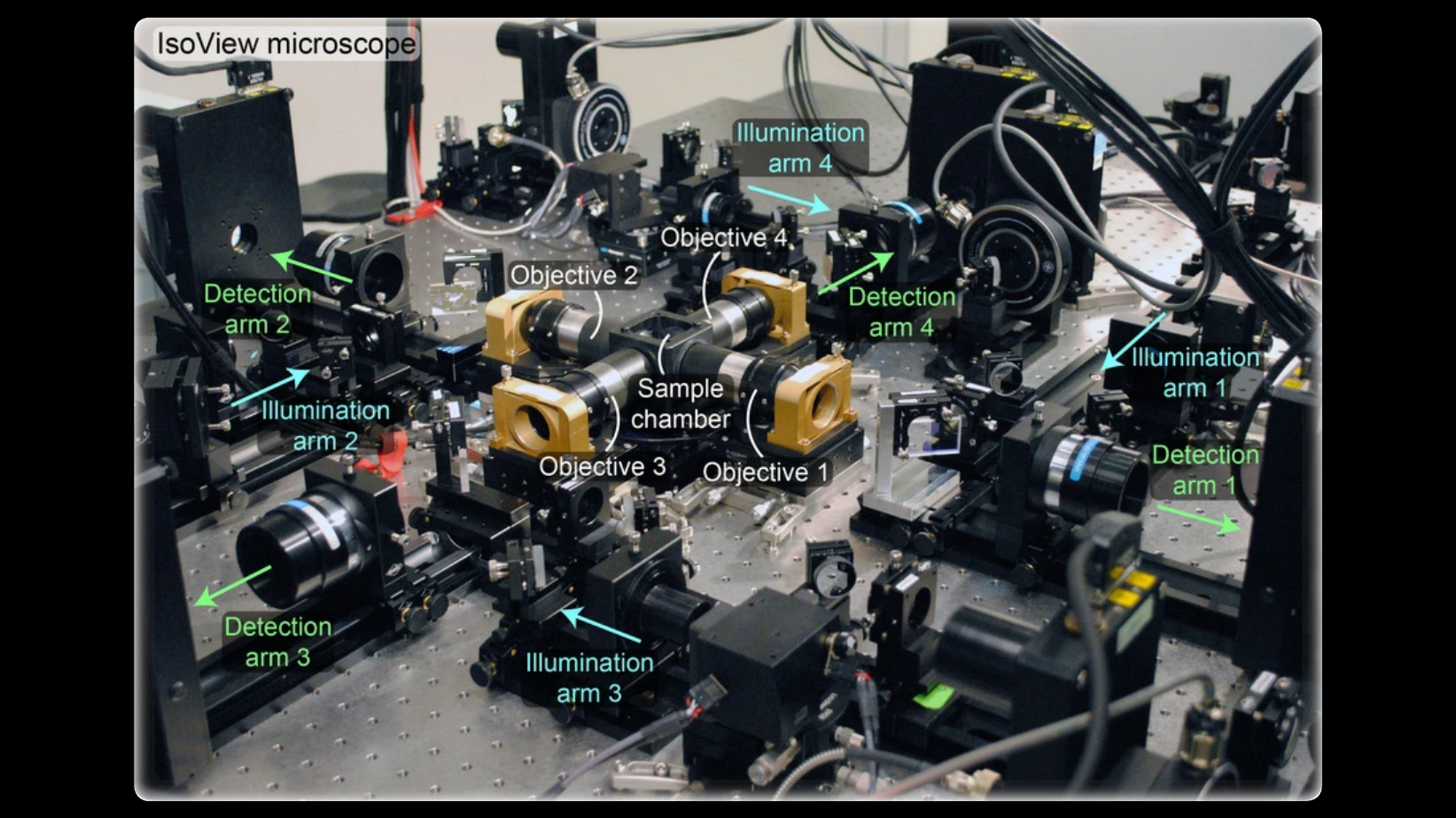
Imaging fast cellular dynamics across large specimens requires high resolution in all dimensions, high imaging speeds, good physical coverage and low photo-damage. IsoView (Isotropic multiview light-sheet microscopy), [1] originally developed at HHMI Janelia Research Campus, achieves these requirements by rapidly imaging large specimens via simultaneous light-sheet illumination and fluorescence detection along four orthogonal directions.
The IsoView design offers volumetric imaging speeds of up to 20 Hz, an isotropic spatial resolution of 400 nm, and maximizes physical coverage by capturing multiple views of the specimen. The design facilitates minimally invasive in vivo imaging of embryos and larvae from hours to days without sacrificing temporal and spatial resolution.
IsoView is a next-generation extension of high-speed SiMView (Simultaneous multiview light-sheet microscope), [2] a class of light-sheet microscope that utilizes digitally-scanned light-sheets and fast volumetric acquisition to capture opposing views of a specimen simultaneously. The SiMView framework has enabled in vivo imaging of entire Drosophila, zebrafish, and mouse embryos for automated tracking throughout embryogenesis. As a superset, unique capabilities and features of SiMView, such as the AutoPilot framework [3] to continuously adapt the microscope to spatiotemporally varying optical properties of the specimen, are directly translatable to IsoView.
References
[1] Chhetri et al., Nature Methods (2015)
[2] Lemon et al., Nature Communications (2015)
[3] Royer et al., Nature Biotechnology (2016)

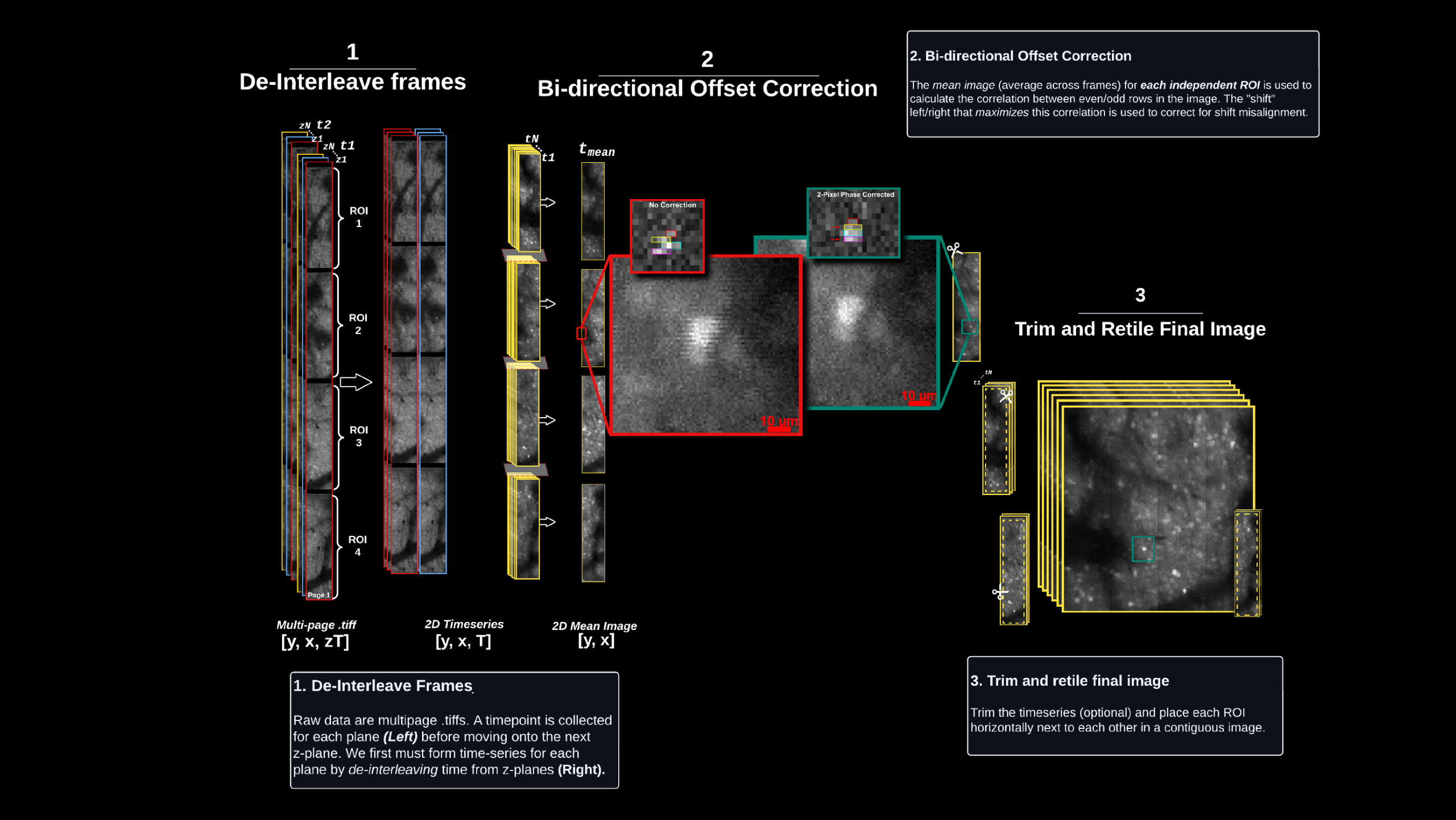
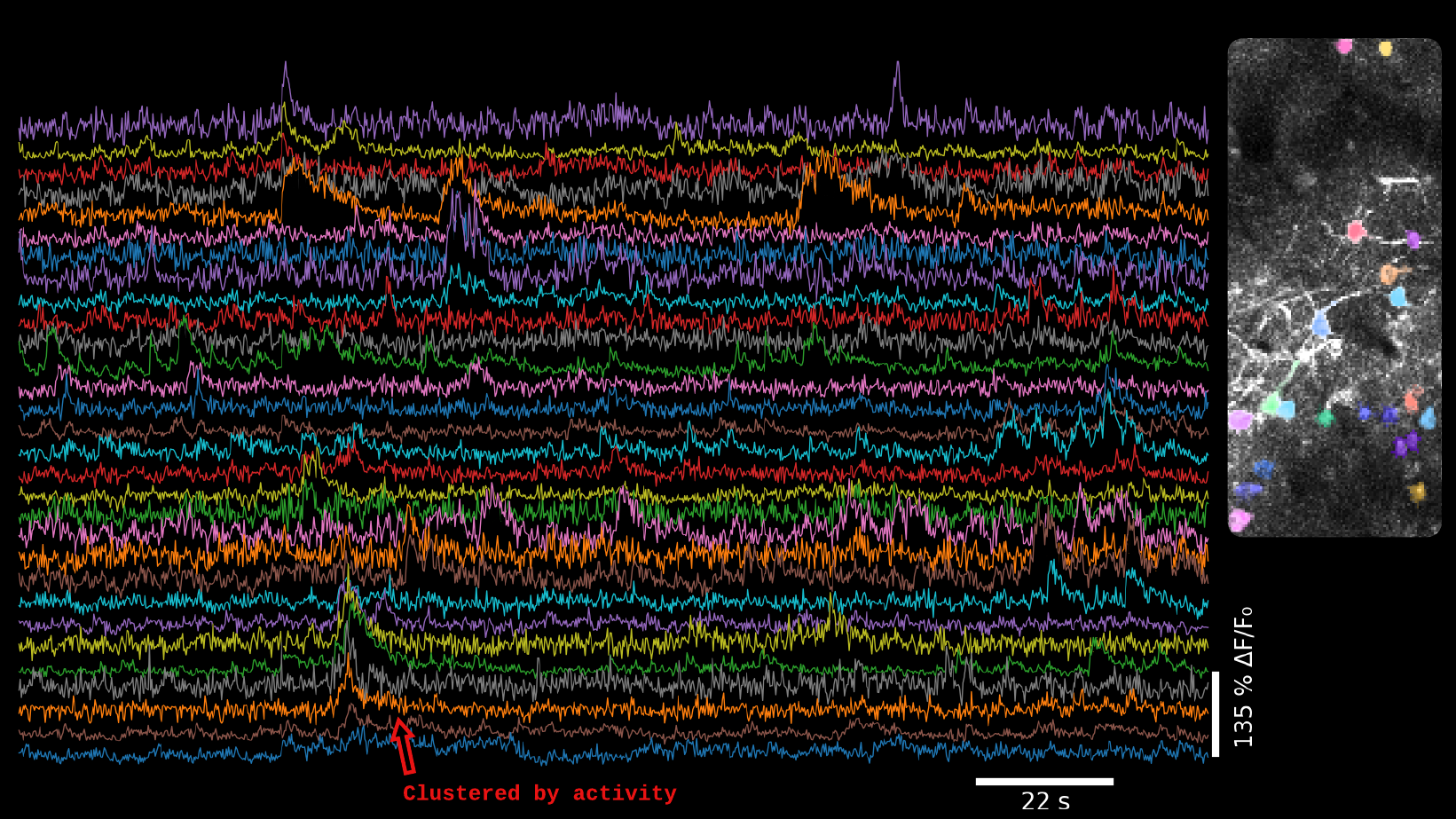
Computational pipeline to extract, motion correct, segment and extract functional activity in LBM datasets leveraging the MATLAB implementation of CaImAn [4].

Computational pipeline to extract, motion correct, segment and extract functional activity LBM datasets leveraging the PYTHON implementation of CaImAn [4].
- A reconfigurable LBM module to seamlessly switch between cellular resolution and sub-micron resolution LBM imaging
- Extension of IsoView light-sheet microscopy to image a variety of cleared tissue samples